<Figure1>
Research team in the graduate school of science and technology (left), and research team in the graduate school of medicine (right).
HOME > Research and Development Projects Adopted in FY2015 > Development of Core Technology for Therapeutic Xeno Nucleic Acid Aptamers
Project Leader:Kuwahara Masayasu
Nihon University, College of Humanities and Sciences(2018/04-)
Nucleic acid aptamers are single-stranded DNA/RNA molecules that can specifically bind to their targets, similar to the function of antibodies; they are expected to be applied to therapeutic drugs and diagnostic agents. Recently, technologies for biomarker discovery in our country have reached the top international level; various biomarkers related to diseases, such as cancers and various circulatory system diseases, are being discovered. Furthermore, owing to Asian economic growth and globalization, MERS coronavirus, dengue virus, SARS coronavirus, pollutants containing PM2.5, and foods containing toxic substances pose substantial threats to public health. Therefore, social needs for methods to specifically detect and inhibit these targets have arisen; technological innovations that enable the creation of various artificial receptors specific to emerging targets in a timely manner have become a pressing need. Antibodies are a representative example but are expensive to develop and manufacture. By contrast, nucleic acid aptamers can be provided at a relatively low cost; in addition, the drastic conformational changes that can result when nucleic acid aptamers bind to targets are expected to be applicable for designing biosensors with unique signaling mechanisms that differ from those of antibodies. Furthermore, in the SELEX selection of nucleic acid aptamers, cells and tissues themselves can be treated as targets even though the target molecule is not identified from such components.
In this study, we are attempting to establish a core technology for therapeutic nucleic acid aptamers containing locked nucleic acids (LNAs), which are known to exhibit enhanced biostability, that is, nuclease resistance. Furthermore, we will combine those xeno nucleotides with base-modified nucleotides to expand the structural diversity of nucleic acid aptamers. As an example, we will develop therapeutic xeno nucleic acid aptamers for adrenal adenoma and disease markers of aldosteronism. This project is promoted by the collaboration of the Graduate School of Science and Technology and the Graduate School of Medicine including university hospital. Furthermore, Prof. Obika�fs group at the graduate school of pharmaceutical sciences at Osaka University; National Institutes of Biomedical Innovation, Health and Nutrition (NIBIOHN) will be technically assisting this project.

Research team in the graduate school of science and technology (left), and research team in the graduate school of medicine (right).

Capillary electrophoresis apparatus for the CE-SELEX method; this method enables simple and quick acquisition of nucleic acid aptamers because of the ability of CE to separate sequences with high avidity from inactive sequences. Further advantages of this method include automated operation from sample injection to the fraction collection process and no requirement to immobilize targets on a solid phase, which is an essential step in the conventional SELEX method.
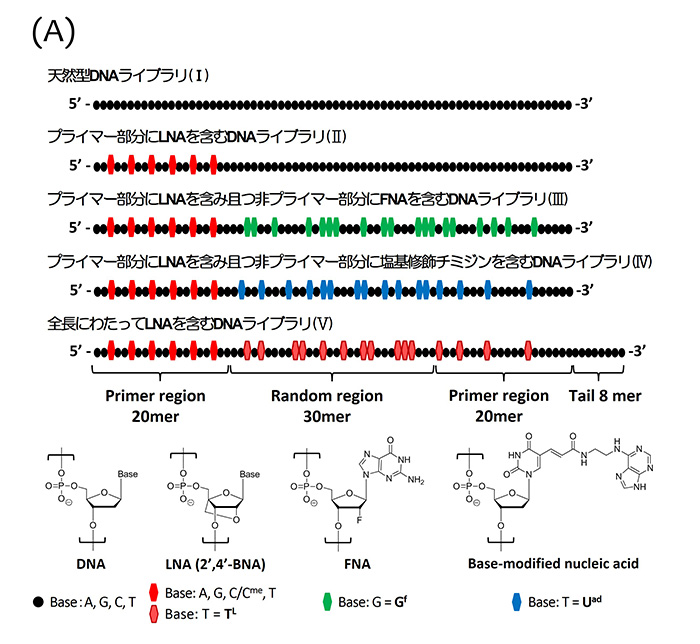
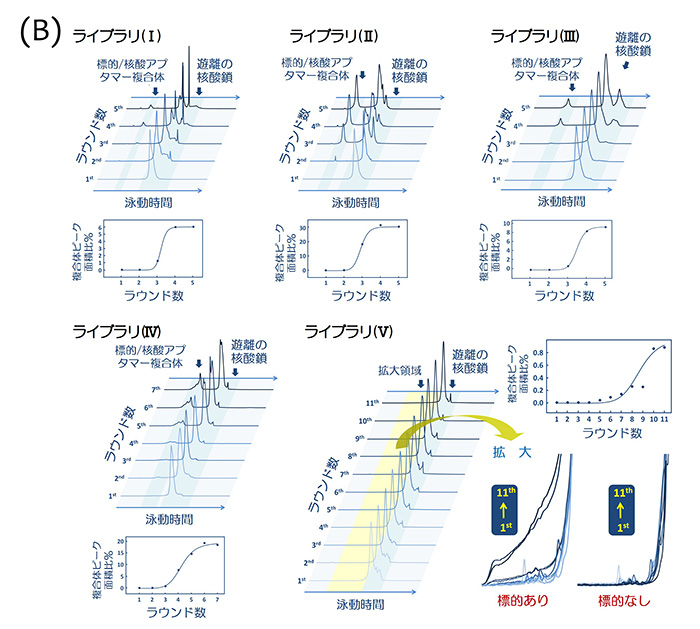
Selection of nucleic acid aptamers by CE-SELEX; (A) one natural DNA library (I) and four chemically modified DNA libraries containing LNA (II?V); (B) progress in the enrichment of each library. We first obtained chemically modified DNA aptamers containing LNA nucleotides in both primer and random regions from the library (V).