Research and Development Projects Adopted in FY2014
Acceleration of the development for the next generation bio-drugs based on macrocyclic nonstandard peptides
Project Leader:Suga Hiroaki
Professor, Graduate School of Science, The University of Tokyo
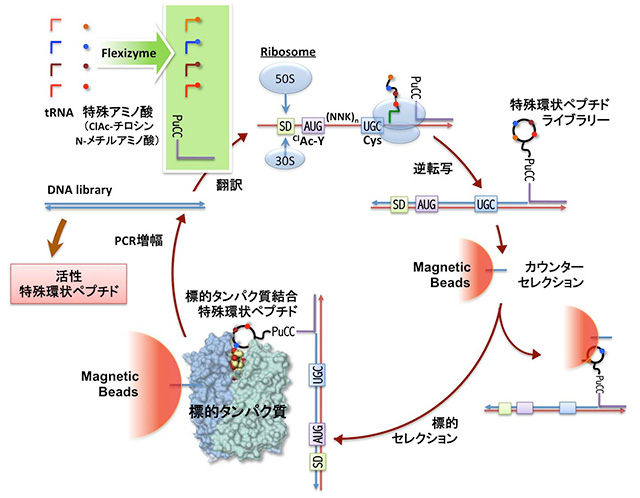
Suga et al. have developed a breakthrough drug discovery platform for natural product-inspired peptides, referred to as RaPID (Random non-standard peptide integrated discovery) system (Fig. 1).
In this system, the flexizyme technology, consisting of a set of flexible tRNA acylation ribozymes, is integrated with mRNA display expressed by a reconstituted in-vitro translation system, referred to as FIT (Flexible In-vitro Translation) system.
This system enables us to readily prepare various libraries of natural product inspired macrocyclic peptides and rapidly screen highly potent ligands again various target proteins in an inexpensive manner.
The "Screening" in the RaPID system is distinct from the classical screening of compounds library, which involves expression of the individual macrocyclic peptides that covalently fused to the respective mRNA templates.
The active species of macrocyclic peptides will be then isolated by the affinity selection to the target protein immobilized on magnetic beads. The mRNA sequence of the isolated peptide-mRNA fusion was reverse-transcribed into cDNA,
followed by PCR amplification to enrich the active DNA template sequence. By means of this system, we are able to "select" potent ligands against a target of choice from the library consisting of a trillion members of macrocyclic peptides expressed from the respective mRNA templates.
Several selection rounds (usually 6 rounds) yield highly potent inhibitors with dissociation constants ranging from single digit nM to sub nM within a few weeks.
We have already succeeded in discovering such macrocycle ligands against over 30 different kinds of targets with the success rate of above 90%.
It should be noted that the molecular weights of the macrocyclic peptides are ranging from 2,000 to 3,000 Da, which are far smaller than antibodies, and thereby they have potential to passively penetrate into cells, i.e. they can target to intracellular proteins.
We believe that this technology is a powerful, reliable, and unique drug discovery platform from Japan.
We thus plan to use this RaPID system to generate novel second generation bio-drugs and revolutionize the drug discovery and development. We set three specific aims in this project.
- Specific aim(1):Revolutionized macrocyclic peptide-based delivery vehicles specific to disease cells
- We will construct cPDC (cyclic Peptide-Drug Conjugate) that is able to specifically deliver an appropriate drug to disease cells.
- Specific aim(2):Revolutionized mid-sized bio-drugs based on macrocyclic scaffolds
- We will develop a discovery platform for cell membrane-permeable macrocyclic peptides capable of effectively inhibiting protein-protein interactions.
- Specific aim(3):Revolutionized therapeutic applications
- We will build our knowledge and experiences in the above drug leads for the therapeutic applications.

<Figure1> RaPID System
The technology, which is the original technology developed by Suga, is built by the combination of ribosomal synthesis of macrocyclic nonstandard peptide libraries with mRNA-display,
enabling us to enrich active species that bind to a target protein, the recovery of cDNAs pairing with their mRNAs, and PCR amplification of active template sequences.
Moreover, the collection of the DNA sequences are transcribed to mRNAs, which are translated to form the peptide-mRNA fusions for repeating the above cycles.
The N-chloroacetyl-Tyrosine (ClAc-Y) group introduced at the N-terminus of peptides spontaneously reacts with a cysteine residue at a downstream position of the random peptide sequences to form macrocyclic peptides via a non-reductive thioether bond.
SD, ribosome binding site; n of (NNK) n is varied from 8 to 15, and their mixture; N of (NNK)n is the mixture of UAGC bases, K is the mixture of U or G; (GGC AGC)3 was translated to a (GC)3-peptide linker.