Development of the technology for synthesizing VHH antibodies with high affinity and stability based on a novel repertoire of genetically encoded amino acids
Project Leader:Sakamoto Kensaku
Group Director, Bio-Functional Molecule Development Group, Division of Structural and Synthetic Biology, Center for Life Science Technologies, RIKEN
A VHH antibody is a single-domain protein ten times smaller than IgG, consisting of the antigen-binding domain of a certain-type of camelid antibody.
It has three loops involved in the binding [complementarity determining region (CDR)], like an H chain of IgG.
With no sugar chain attached, VHH antibodies can be synthesized by bacteria.
These features make it a lot easier to produce VHH antibodies and maintain their quality, as compared with IgG, which means huge potential of industrial application.
Its single-domain composition probably helps VHH to avoid the formation of unsolvable aggregation, which is a property associated with antibodies consisting multiple domains, and to maintain its activity under harsh conditions.
Thus, VHH might be applied to medical and diagnostic areas that are not suitable for other antibodies.
VHH antibodies have been obtained by immunizing camels or alpacas and then screening useful clones using the phage-display technique.
However, the immunization of camelid animals takes a lot of time, and the in vivo process of maturing antibodies is so invisible as to cause a concern that there is some limitation on the diversity of obtained antibodies.
These concerns have prompted the efforts to isolate VHH antibodies from artificial random libraries, but it is still difficult to obtain VHH antibodies matching those obtained by immunization in quality, such as the affinity for antigens and the structural stability.
VHH is among the proposed protein formats to replace IgG in industry, such as various types of antibody fragments, conjugates between antibodies and other proteins, and artificial protein scaffolds.
Nevertheless, we stick with VHH and have decided to develop technologies to facilitate its industrial use, because VHH must have a good “inborn Equality, being derived from natural antibody and consisting of a single domain.
We envisioned that the affinity and stability of VHH antibodies might be improved using an expanded repertoire of amino acids, as we possess a cell-based platform for incorporating designer or non-natural amino acids into proteins.
This in vivo technology is an outcome of the many-year quest for “artificial genetic codes Eby Dr. Sakamoto and his colleagues.
In May, 2015, Dr. Sakamoto’s group and a research team in the US independently reported that the expanded amino-acid repertoire will provide unique solutions to many of the problems of protein engineering.
In his report, Dr. Sakamoto indicated a facile way to enhance the structural stability of proteins by incorporating a non-natural amino acid at multiple sites.
We intend to develop methods to improve the affinity and stability of VHH antibodies, exploiting the non-natural amino acid platform.
The University of Tokyo (represented by Dr. Satoru Nagatoishi) and Ajinomoto Co. Ltd. take part in the present project commissioned to RIKEN.
Dr. Nagatoishi is an expert on analytical methods and physico-chemical aspects of the interaction between antibodies and antigens.
The goal of this project is more than developing individual VHH antibodies; the platform developed through this project will cater to pharmaceutical and other industries, with Ajinomoto playing a decisive role here.
<Figure1>
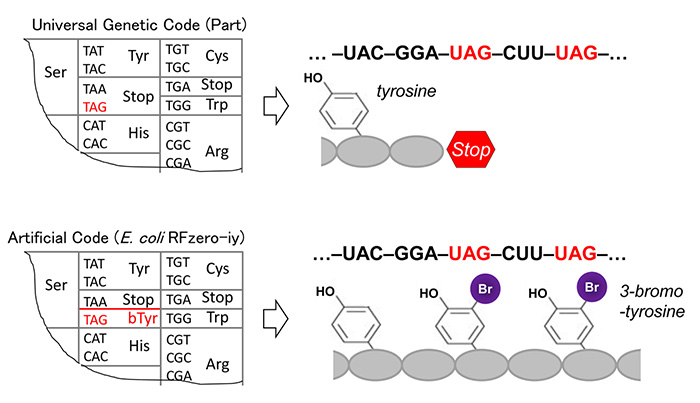
Recombinant protein synthesis using artificial genetic codes.
The UAG codon is a stop codon in the standard code, and a given base sequence is translated accordingly (Upper panel).
In E. coli RFzero-iy strain, its meaning has been change to mean a halogenated tyrosine, and proteins will contain this synthetic molecule at UAG position (Lower panel).
<Figure2>
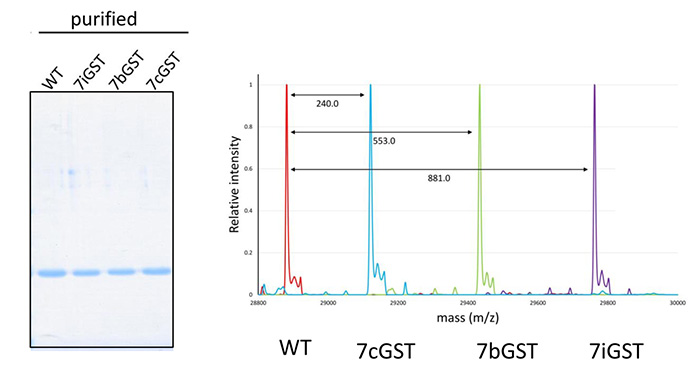
Yields and homogeneity of recombinant proteins containing designer amino acids. The yield after purification was almost the same between the proteins with halogenated tyrosines at 7 positions and the wild-type molecule (with no halogenated tyrosine) (Left panel). Mass spectrometric analyses showed the homogeneity of each of the halogenated proteins, the masses of which were each larger than that of WT by the total mass of seven halogen atoms (Right panel).
<Figure3>
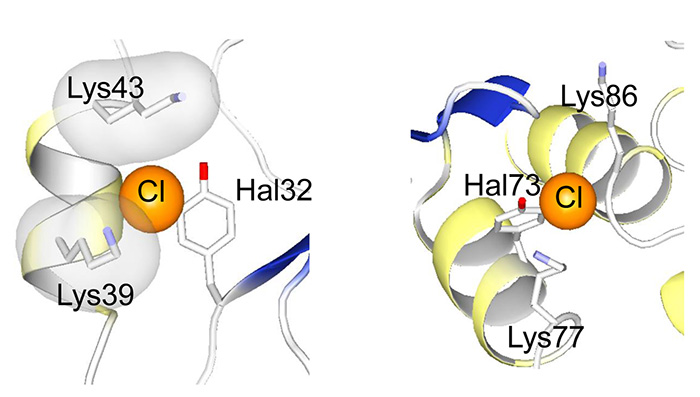
How chlorotyrsines are incorporated into a protein molecule. An X-ray crystallography indicated the local structures around the incorporated halogenated tyrosines. The halogen atoms attached to the tyrosine rings fill the internal space of the protein molecule and thus create new interactions with neighboring residues.
<Figure4>
Two colleagues from Ajinomoto Co. Ltd. standing before a panel showing the company’s policy.